Using bibliometric techniques to assess the state of scientific literature on COVID-19
Like so many other scientists during the first year of COVID-19, I was struck by the research response to the pandemic and the resulting information overload for scientists. How can we make sense of all this information? Nguyen Tran, a doctoral student who I co-advise, and I decided to try to tackle this problem using a method common in library and information sciences: bibliometric analysis. By applying bibliometric techniques, we were able to bring perspective to some of the early literature on COVID-19. This blog post is a rough sketch of an article we intended to publish, but unfortunately the data we were working with became dated pretty quickly as the pandemic evolved in both size and scope. As such, Nguyen and I decided to share the manuscript on this blog as a way to openly document our methods in the hope that it will benefit other epidemiologists who may be interested in this technique. I think bibliometrics has unique applications in our field, for example, making sense of free text data in the electronic health record where each patient encounter could be conceived as the literature for a patient.
Abstract
Researchers and scientists have responded in force to the COVID-19 pandemic of 2019-20. Much of this scientific information has been shared ahead of print via preprint servers. In order to help shed light on research priorities and trends, and guide researchers in gaps in the literature, we performed a bibliometric analysis of preprint articles pertaining to COVID-19 disseminated via bioRxiv and medRxiv (n=1,890 manuscripts as of April 18, 2020). We conducted two main analyses of these data: research themes by time and research productivity by geography. In an exploratory aim, we constructed hierarchical predictive models for repeated measures to assess whether the ecological drivers of research output were a function of the COIVD-19 pandemic or a by-product of other country-specific factors. Our thematic analysis demonstrated a clear trend in the research activity, from pathogenesis, to outbreak epidemiology, and most recently clinical management and public health prevention. The majority of manuscripts were from China and the United States (55% of manuscripts), followed by the United Kingdom, India, Italy, and Germany (20% of manuscripts). In the predictive model, research output appears to be driven by time and the number of universities in a given country, as opposed to the number of confirmed cases. For research topics that are saturated, this may suggest that meta-analysis or systematic reviews are needed to synthesize existing evidence. For research topics that are not as well studied, this may motivate grant proposals, prospective research projects, and secondary data analysis of un- or under-utilized data.
Introduction
The unprecedent COVID-19 pandemic of 2019-20 has generated an equally unprecedented response from the scientific and academic community in the form of data and research. The resulting information overload presents a challenge for those responding to the pandemic, and it may be difficult to ascertain the appropriate research needed at a given time during the pandemic. The question arises, does the research itself shed light on research priorities and trends, and can this help inform other researchers of gaps in knowledge? This may be answerable by examining trends in research dissemination through bibliometrics (1).
Given the urgency of the pandemic, scholarly research is being rapidly disseminated through what are known as preprint servers. arXiv (https://arxiv.org), which is one of the most popular and well known (2), allow researchers to share their findings with others prior to peer review and represent an intermediary product in the publication process (3). Such servers are being extensively utilized for COVID-19 relevant research. Indeed, inspired by arXiv and specific for research in the biomedical domain, bioRxiv (https://www.biorxiv.org) and medRxiv (https://www.medrxiv.org) have separate parts of their website dedicated to COVID-19 manuscripts (4). As formal peer review may delay dissemination of important findings when responding to an emergency, such as a pandemic, we contend that these preprint servers are a valuable source of information on the state of COVID-19 research.
Our primary aim was to describe the state of research on COVID-19 disseminated preprint to identify research trends. In order to ground our research and make our aim tractable, we focused on two specific attributes of research dissemination: time and geography. First, by exploring research dissemination as a function of time, we hope to depict the evolution of research projects and priorities as an indicator the state of scientific knowledge. Second, by exploring dissemination as a function of geography, we sought to identify the locales that are contributing the preponderance of scientific research. As a secondary exploratory aim, we further describe time and geography alongside pandemic surveillance data and develop a predictive model of COVID-19 research output. Our overall hypothesis is that the declaration of a pandemic has been a "call to arms" among researchers, scientists, and lay citizens worldwide and we would expect to see non-peer reviewed research output mirror the trajectory and geography of the pandemic itself.
Methods
To ascertain the state of research emerging real-time during the pandemic, we focused on publications identified as COVID-19 on two preprint servers: bioRxiv and medRxiv (4). On April 18, 2020, we downloaded and imported the manuscript metadata including title, date, author list, and abstract for our analysis. As motivated earlier for our primary aim, we conducted two main analyses of these data: research by time and research by geography. Data corresponding to COVID-19 confirmed cases obtained from the World Health Organization (WHO) were linked to the manuscript data for our exploratory secondary aim (5).
In the "by time" analysis, manuscripts were aggregated on a per week basis and analyzed using the tidytext R package (6). This package was used as an automated method for text mining and natural language processing given the large number of abstracts. Briefly, the algorithm analyzed the text of the manuscripts' titles and abstracts, and constructed measures of term frequency and inverse document frequency, a weighting approach that down-weights frequently appearing terms and up-weights infrequently appearing terms. Based on the frequency of identified keywords for each week generated via the automated approach, we inductively assigned themes as a proxy for the type and focus of the research. These data were then plotted and compared to WHO COVID-19 confirmed case data in order to depict the time trend as it related to reported cases adjusted for population density (people/km2).
In the "by geography" analysis, the first author of each manuscript was geocoded to their respective country according to their primary institution; when the first author institution was unavailable or failed to geocode, we used the last author institution as a substitute. These data were plotted on a global map and again compared to the WHO COVID-19 confirmed case data to assess whether countries with a higher number of cases were associated with a greater number of preprint manuscripts. We used choropleth maps with additional symbology as a way to depict both variables (case count and manuscript count) simultaneously on a single map projection. As operationalized in the map, the color gradient reflected the population density adjusted number of confirmed cases, and the size of the symbol within each country depicted the crude number of preprint manuscripts.
To identify the principal drivers of research output during COVID-19, and whether these main drivers were a function of the pandemic itself or a by-product relating to other country-specific factors, we constructed hierarchical negative binomial models for repeated measures. These models assessed the ecological association between the number of manuscripts per population density (dependent variable) and the independent variables: the number of reported cases per population density, a continuous measure of time, and number of universities. Both the number of reported cases and the number of universities were standardized as Z-scores to facilitate modeling and interpretation. We also hypothesized that there was likely a time-lag between the reports of confirmed cases and research output; thus, we modeled a 2-week lag such that for the total number of preprint manuscripts in a given week, the total population density adjusted confirmed cases would be from 2 weeks prior. We considered a 1-week lag as part of a sensitivity analysis, assuming that responses to the pandemic may be quicker than anticipated. Random effects of countries on the intercept were modeled to account for country-level differences in the initial number of publications and reported cases of COVID-19. The independent variables were added individually to assess their effect on the dependent variable, and model fit was evaluated using -2 times the change in log-likelihood, where lower values represented a better overall fit to the data. All analyses were conducted in R version 3.6.3, and analytic code and data are available for download from https://doi.org/10.5281/zenodo.3773809.
Results
As of April 18, 2020, there were 1,890 manuscripts shared via bioRxiv and medRxiv, and 2,164,078 confirmed cases of COVID-19 reported to the World Health Organization.
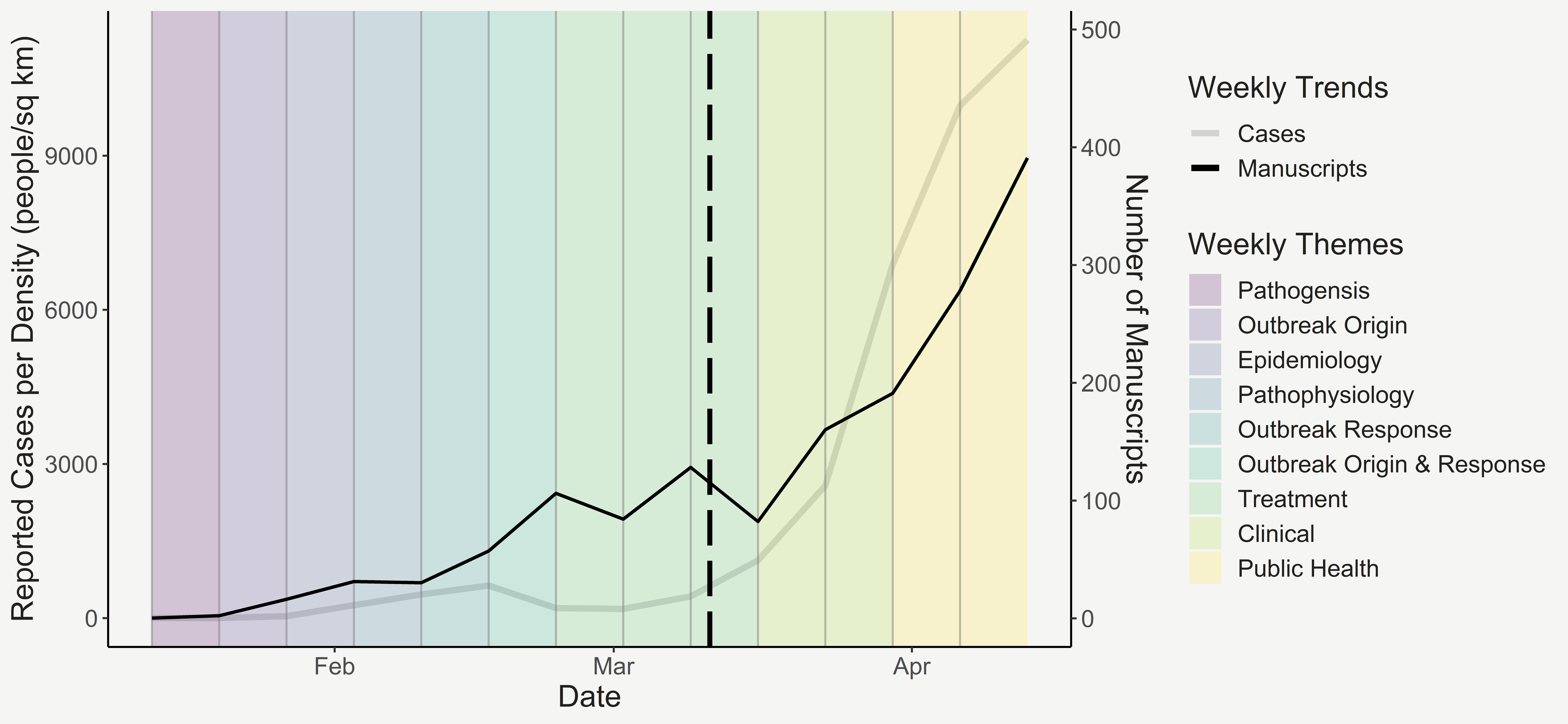
The above figure shows a clear - and unsurprising - increase in both reported cases of COVID-19 (adjusted for population density) and research being posted to the preprint servers. The total numbers increased dramatically after the declaration of a pandemic by the World Health Organization on March 11, 2020. Our thematic analysis demonstrated a trajectory of research that started with the basic sciences of virology and genetic biology in understanding disease pathogenesis and proceeded towards understanding the origins and scope of the outbreak through epidemiological investigations around the time the pandemic was declared. More recent work has focused on clinical aspects of disease including treatment and public health preventive efforts.
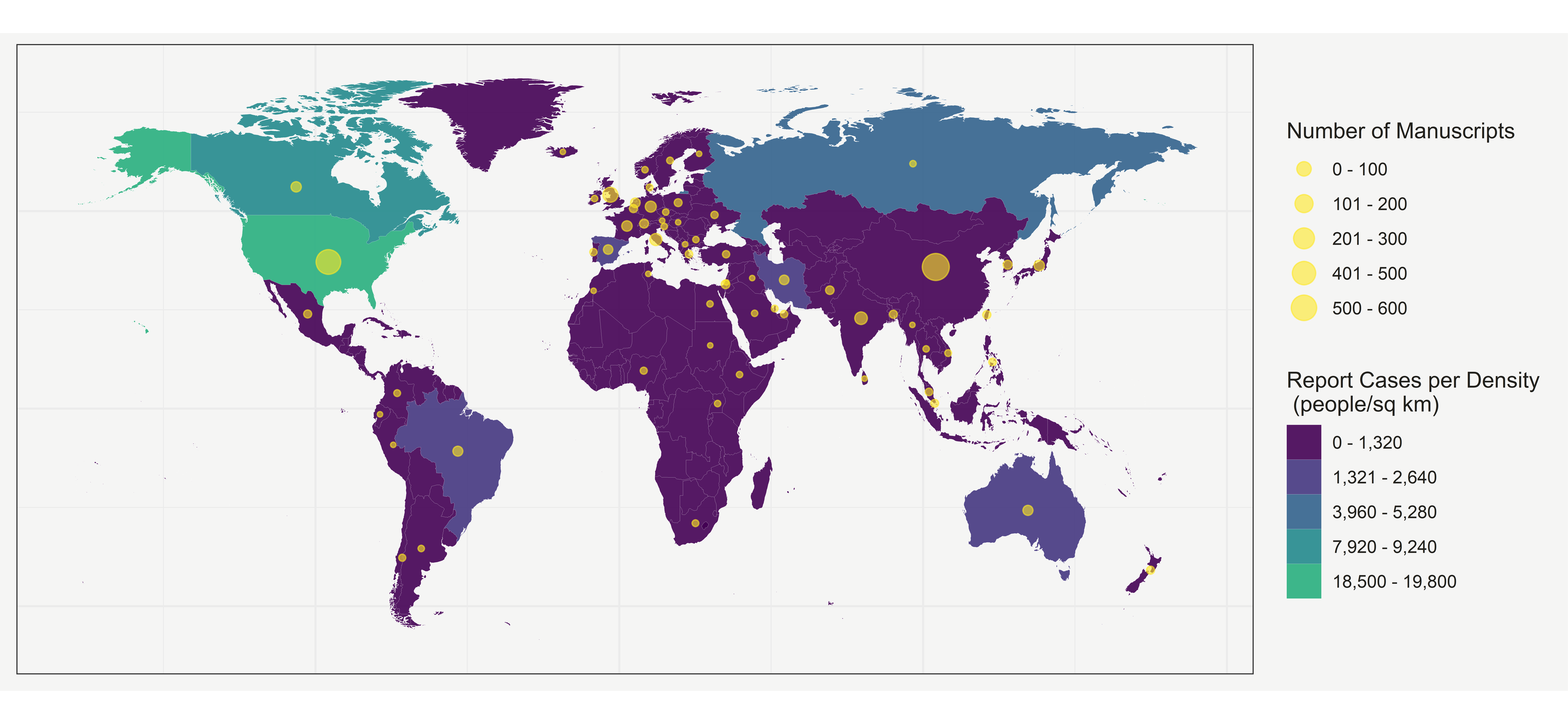
The above figure depicts the geography of confirmed cases of COVID-19, adjusted for population density, and the crude number of preprint manuscripts. The two main focal areas for manuscripts were from first authors affiliating with an institution in China or the United States. Researchers in both of these countries had submitted in excess of 400 manuscripts posted to the preprint servers (n=583 for China, n=462 for the United States). Over 75% of manuscripts was submitted from these two countries, plus the United Kingdom (n=115), India (n=64), Italy (n=56), and Germany (n=40). The video linked here demonstrates the progression of manuscripts over time, and shows China, as the early epicenter, leading the way.
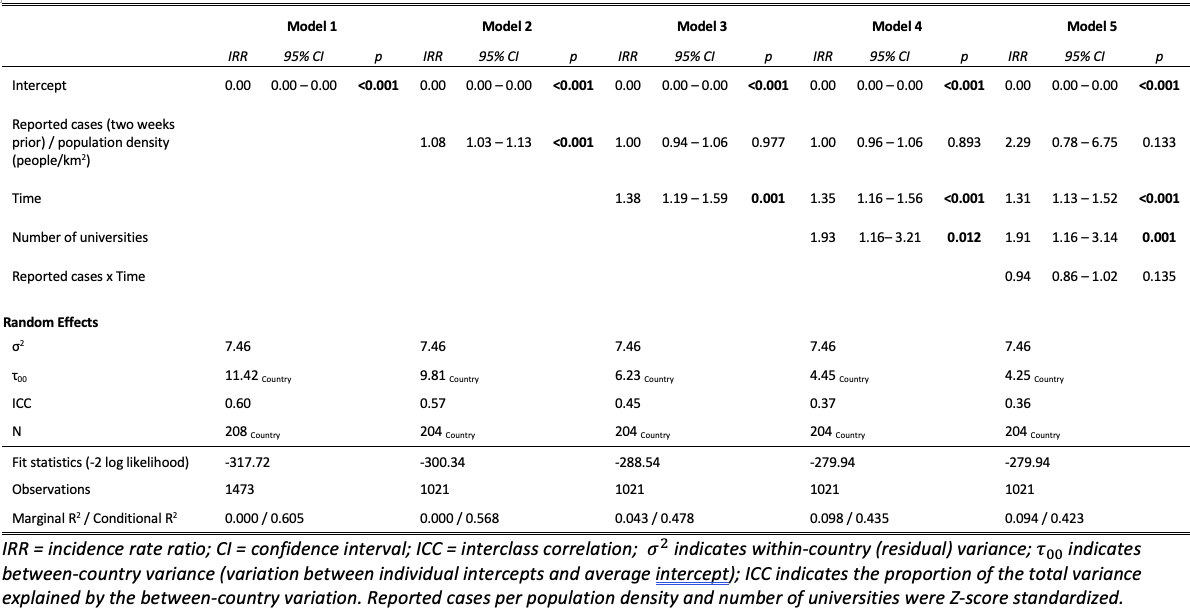
The ecological associations of each independent variable with the population density adjusted number of preprint manuscripts is shown in the table below. Model 1 shows that about 60% of the total variation in the number of manuscripts per population density is explained by between-country differences. When only the effect of confirmed cases per population density was assessed (model 2), there appeared to be a marginal correlation such that a unit increase in the standardize Z-score for the population density adjusted confirmed cases was associated with an 8% increase in the population density adjusted manuscripts (IRR = 1.08; 95% CI: 1.03, 1.13). However, this association did not persist when we also considered the effect of time (model 3) and Z-score standardized number of universities (model 4), both having a positive association with the population density adjusted manuscripts. In our final model, the results did not indicate an interactive effect between time and reported cases per population density on the dependent variable. Our sensitivity analysis also showed that the estimates were robust to the time-lag difference.
Discussion
In this exploratory analysis of manuscripts posted to two preprint servers early in the COVID-19 pandemic, we observed a strong global response from the research community in line with the evolution of the pandemic. In countries with some of the higher number of confirmed cases per population density, we also found the highest number of preprint articles. However and contrary to our hypothesis, the predictive model suggested that there may be other country-specific factors that are the primary drivers of the growing number preprint publications as opposed to the density of cases. Indeed, in many countries affected by COVID-19, there has been little to no preprint research disseminated on bioRxiv and medRxiv: most of these publications have been from researchers affiliating with institutions in the United States. This may reflect the English-language and U.S.-based bias of these servers. While publication of scientific findings lean towards English language-based journals (7), this nevertheless paints an incomplete picture of the state of COVID-19 research, particularly is it may apply to more nuanced or locale-specific data.
Our thematic analysis results can be used by researchers to assess priorities and gaps in the literature as follows. For research topics that are saturated, this may suggest that meta-analysis or systematic reviews are needed to synthesize existing evidence. For research topics that are not as well studied, this may motivate grant proposals, prospective research projects, and secondary data analysis of un- or under-utilized data. Specific to data scientists, our thematic analysis findings underscore the importance and opportunity for collaboration across a host of disciplines: from basic sciences such as virology and immunology, to the social sciences of public health and policy, to the health sciences of clinical epidemiology and medicine.
Despite our focus on bioRxiv and medRxiv there are, of course, other sources of research related to the pandemic, including other preprint servers, such as the Open Science Foundation (https://osf.io/preprints/) and Zenodo (https://www.zenodo.org/communities/covid-19/), as well as traditional peer-reviewed journals. One platform is even attempting to catalog all research posted, whether peer-reviewed or preprint, or in a conference proceeding, and as of this writing there were close to 10,000 COVID-19 related publications (8). If, in fact, the manuscripts posted to bioRxiv and medRxiv are representative of the greater body of research, our findings are particularly useful to shed light on what research has been conducted, and potentially, gaps in the knowledge corresponding to research needed. For non-English speaking audiences, there is even a tool that translates preprint results to over 100 other languages. Nevertheless, this does not take the other direction into account - retrieve non-English based articles through the English-based preprint servers - and therefore is still an incomplete view.
The use of bibliometrics to evaluate the products of COVID-19 research is a growing area. The National Library of Medicine has their own clearinghouse of over 7,500 publications and provide utilities to describe trends (9), and we have even noted a dedicated website cataloging and analyzing COVID-19 publications (https://www.covid19bibliometrics.org). Most of these other resources are based on articles published in the peer-reviewed literature. A small body of other COVID-19 bibliometric analyses have been limited to peer-review publications alone, often based on manual searches (10-12). Our work builds upon these efforts by focusing on the rapid dissemination of articles preceding publication, and thus reflects a much wider body of scientific information and may be the most comprehensive analysis as of this writing. Encouragingly, our findings largely mirror these early efforts.
We wish to caution readers about overinterpreting bibliometric COVID-19 studies, ours included, for several reasons. First, when relying upon preprint manuscripts, the quality of the research is unknown. Indeed, our sources even include a strongly worded disclaimer that cautions readers the preprint manuscripts "should not be regarded as conclusive, guide clinical practice/health-related behavior, or be reported in news media as established information." Second, while some countries appear to have a greater population density adjusted number of COVID-19 cases, this should not be conflated with prevalence or true disease burden as differences may reflect patterns of testing and reporting. Third, the number of manuscripts being produced by authors affiliating with institutions in specific countries may in fact relate to underlying academic productivity in these areas, not specifically a pandemic response. Relatedly, our ecological predictive model is not a causal model. That is, we are not attempting to suggest that the burden of pandemic resulted in disproportionate manuscript dissemination as there may be myriad reasons for producing academic output, as discussed earlier. Rather, our interest was to identify patterns and generate hypotheses that can then be thoroughly vetted in subsequent work. Bibliometric analyses may not reveal causal mechanisms but demonstrate visually the research community's response to the pandemic. Strengths of our work include focusing on preprint, not peer reviewed, manuscripts, the use of automated text mining approaches to process a large number of articles, and informative visualizations.
In short, for COVID-19 researchers to understand their next steps, it is useful to visit not only published work - a traditional literature review - but also the larger body of unpublished work seeking to understand the pandemic.
The authors thank Daniel Vader of the Drexel Dornsife School of Public Health and Dnyanada Kadav and Steven Melly of the Drexel Urban Health Collaborative for technical support.References
- Hand E. Researchers aim to chart intellectual trends in arXiv. Nature. 24 February 2012. doi:10.1038/nature.2012.10103
- Van Noorden. The arXiv preprint server hits 1 million articles. Nature. 30 December 2014. doi:10.1038/nature.2014.16643.
- PNAS policy on prior publication. Proc Natl Acad Sci U S A. 1999 Apr 13;96(8):4215.
- medRxiv. COVID-19 SARS-CoV-2 preprints from medRxiv and bioRxiv. Available at: https://connect.medrxiv.org/relate/content/181. Accessed April 18, 2020.
- World Health Organization. Coronavirus (COVID-19). Available at: https://who.sprinklr.com. Accessed April 18, 2020.
- Silge J, Robinson D (2016). “tidytext: Text Mining and Analysis Using Tidy Data Principles in R.” JOSS, 1(3). doi: 10.21105/joss.00037, http://dx.doi.org/10.21105/joss.00037.
- Amano T, González-Varo JP, Sutherland WJ. Languages Are Still a Major Barrier to Global Science. PLoS Biol. 2016;14(12):e2000933. Published 2016 Dec 29.
- Publons. COVID-19 related publications. Available at: https://publons.com/publon/covid-19/?sort_by=date. Accessed April 18, 2020.
- National Library of Medicine. LitCovid. Available at: https://www.ncbi.nlm.nih.gov/research/coronavirus/. Accessed April 18, 2020.
- Bonilla-Aldana DK, Quintero-Rada K, Montoya-Posada JP, Ramírez-Ocampo S, Paniz-Mondolfi A, Rabaan AA, Sah R, Rodríguez-Morales AJ. SARS-CoV, MERS-CoV and now the 2019-novel CoV: Have we investigated enough about coronaviruses? - A bibliometric analysis. Travel Med Infect Dis. 2020 Jan - Feb;33:101566.
- Lou J, Tian SJ, Niu SM, Kang XQ, Lian HX, Zhang LX, Zhang JJ. Coronavirus disease 2019: a bibliometric analysis and review. Eur Rev Med Pharmacol Sci. 2020 Mar;24(6):3411-3421.
- Chahrour M, Assi S, Bejjani M, Nasrallah AA, Salhab H, Fares M, Khachfe HH. A Bibliometric Analysis of COVID-19 Research Activity: A Call for Increased Output. Cureus. 2020 Mar 21;12(3):e7357.
Cite: Goldstein ND. Using bibliometric techniques to assess the state of scientific literature on COVID-19. Sep 13, 2021. DOI: 10.17918/goldsteinepi.