Machine learning in electronic health record research
Note: This blog post is an excerpt from the forthcoming book A Researcher's Guide to Using Electronic Health Records, From Planning to Presentation, 2nd edition.
There are at least four areas that machine learning (ML) can contribute to electronic health record (EHR) research: 1) exploratory analysis of large datasets through unsupervised learning, 2) development of predictive models, and to a limited extent, etiologic models through supervised learning, 3) handling missing data, de-identifying data, and creating synthetic data through semi-supervised learning, and 4) knowledge discovery and data mining. There are a plethora of ML algorithms and selecting the appropriate one depends on the research question, data type, hypothesized effects, interpretability, and flexibility (see James et al., 2021 for a comprehensive list of such algorithms).

Exploratory analysis
One of the most straightforward uses of ML is to identify patterns in data for reducing dimensionality. In an EHR derived dataset, there may be hundreds or thousands of variables available for a given patient, especially if -omics or laboratory measures are available. Most likely, there are many dependencies and correlations across these variables, and therefore the researcher must either choose a subset of variables that are minimally collinear or perform dimension reduction. In a traditional epidemiologic analysis, we might undertake a factor analysis or principal component analysis when the dimension reduction problem occurs independent of the health outcome (i.e., dependent variable). Alternatively, we can employ ML approaches that do not depend on the outcome being present in the data, known as unsupervised learning algorithms.
An unsupervised learning approach seeks to learn about patterns and correlations in the independent variables regardless of the dependent variable. This is especially useful where we have broad clinical states that are heterogeneous in their presentation, and we wish to create latent homogenous groups. In other words, the goal is to identify distinct clinical phenotypes (Wang et al., 2020). Unsupervised learning has been used in EHR data to identify phenotypes of Alzheimer's disease (Alexander et al., 2021) and other ageing-related diseases (Kuan et al., 2021), COVID-19 (Cui, Robins & Finkelstein, 2020), pediatric critical care (Williams, Ghosh & Wetzel, 2018), and mental health (Kung et al., 2021), among others. It also has been applied to aspects of care delivery, such as types of EHR users (Fong et al., 2022), management of sepsis (Fohner et al., 2019), and neonatal care (Chen et al., 2021).
Modeling
Regression modeling is the backbone of biostatistical techniques used in epidemiology. Regression can be used for predictive modeling to identify protective or risk factors for disease, or etiologic modeling in the hunt for causal agents for the purposes of intervening. In a traditional regression analysis, a dependent variable is regressed on one or more independent variables. Using supervised learning ML algorithms, we may also predict the outcome based upon a high dimensional input. This contrasts with unsupervised learning where the outcome is removed from the data. Supervised learning techniques are used for regression (mostly predictive, but there are emerging causal approaches), classification problems, and estimation.
Unlike traditional regression approaches where the researcher is required to specify the model, ML does not make assumptions about the data generating process, such as the presence of interactions. Thus, predictive ML approaches can detect previously unknown associations. However, any time we fit a model, whether a regression model or ML, we are making a claim about how the dependent and independent variables are related, requiring us to bring our substantive expertise to evaluate the model. For example, tree-based algorithms may be useful for detecting unknown interactions, but ML cannot distinguish if the independent variables are mediators instead of confounders.
Data scientists speak of a pipeline to move from data to an ML model (figure below). This pipeline begins with typical observational epidemiology considerations of applying inclusion and exclusion criteria to arrive at the analytic sample and variable list. Once the ML algorithm is selected, the data are stratified into training and testing datasets, where the training dataset is the initial application of the algorithm to propose a model. In an iterative process, hyperparameters that control the ML algorithm are tuned, the model is tested in the testing dataset, and the output evaluated to select a final model. Alternative ML algorithms may also be considered as part of this process. Once a model is selected, the predictive performance is evaluated through measures of sensitivity, specificity, and predictive values, which are presented in what is known as a confusion matrix.
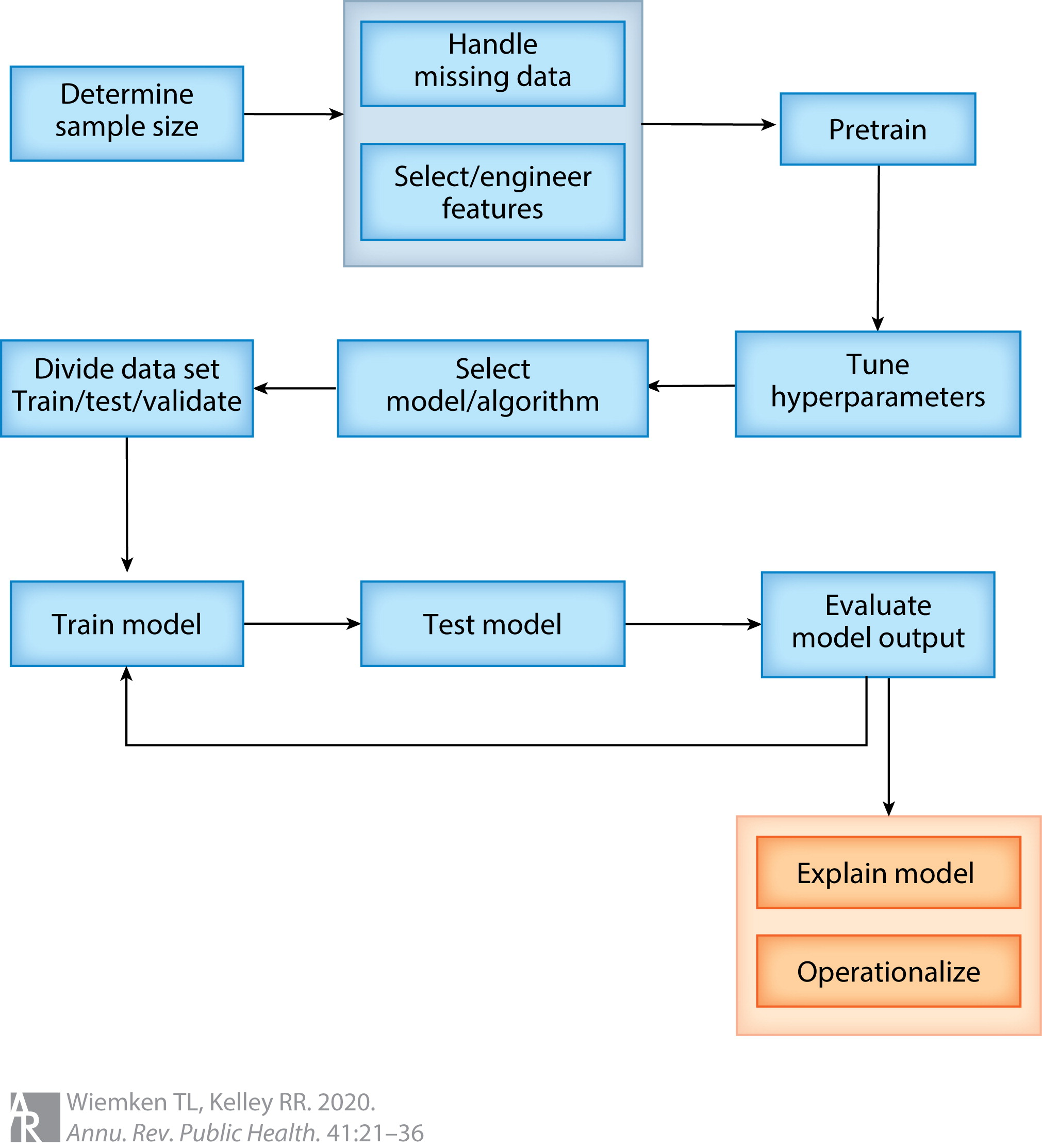
ML-based prediction models have been employed heavily in EHR research. For example, researchers have built models to predict atrial fibrillation (Tiwari et al., 2020), heart failure (Wu, Roy & Stewart, 2010), infection (Bhavani et al., 2020), kidney injury (Kate et al., 2020), post-operative complications (Bronsert et al., 2020), childhood obesity (Pang et al., 2021), diabetes (Dinh et al., 2019), and cardiovascular disease (Dinh et al., 2019); the list goes on extensively. To expound on one study further, Pang et al. developed a "comprehensive machine learning workflow [...] that includes data quality control, data processing, missing data imputation, and machine learning model development for early childhood obesity prediction" using data from a large pediatric EHR. Their ML workflow mirrors the pipeline depicted above. After applying inclusion and exclusion criteria, approximately 27,000 patients with over 100 variables were available for model development., and were divided into training (80%) and testing datasets (20%). Seven ML algorithms, including the XGBoost ensemble algorithm, were developed to predict obesity incidence in the cohort. The authors found the ensemble model performed the best (see Tables 1 and 2 in Peng et al.); unfortunately, a traditional theory driven logistic regression model was not available for comparison. Nevertheless, the authors demonstrated a rigorous process to develop a predictive ML model using EHR data.
Aside from pure prediction, there are emerging approaches to causal modeling using ML. Supervised learning has been used to predict the counterfactual state and fit propensity score models for inverse probability weighting. Through simulation studies, Naimi, Mishler, and Kennedy (2021) contrasted parametric and nonparametric (i.e., ML) based approaches, including inverse probability weighting, g-computation, augmented inverse probability weighting, and targeted minimum loss-based estimation for estimating causal effects. They noted that singly robust ML estimation of the average treatment effect was biased, but performance improved when using doubly robust estimation, where an exposure model and outcome model are combined into a single estimator. The authors offered four suggestions for researchers using ML to quantify the average treatment effect: use 1) doubly robust methods, 2) sample splitting, 3) multiple ML algorithms through an ensemble approach, and 4) interactions and transformations on selected variables.
Under semi-supervised learning, some of the outcome data have been removed. This approach can be used to impute missing outcome data under a variety of missing data mechanisms. Algorithms such as k-nearest neighbors, tree-based methods, and deep learning have embedded capacity to handle missing data as part of the prediction pipeline, potentially enabling its use. In experimental settings, random forest consistently performs well for imputation compared to other more resource-intensive algorithms (Jager, Allhorn & Biebmann, 2021). Despite this integrated capacity, among clinical prediction model studies in the literature, missing data have infrequently been handled through ML (Nijman et al., 2022).
Knowledge discovery and data mining
A third area of ML applications to EHR research is knowledge discovery. In knowledge discovery - also known as data mining - ML provides a method to automatically delve into the EHR and retrieve meaning from the data. A full treatment of knowledge discovery and data mining may be found elsewhere (Freitas, 2002); rather what follows here are applications specific to EHR research.
Knowledge discovery proceeds in a sequential fashion, beginning with data identification, followed by data preprocessing, then data mining, concluding with evaluation. Preprocessing constitutes most of the process (Zhang, Zhang & Yang, 2003). As such, researchers have proposed pipelines for data preprocessing prior to the application of ML techniques: in the pipeline figure, this is an expansion of the second step following sample size determination. One such pipeline is known as FIDDLE (Flexible Data-Driven Pipeline), which "systematically transforms structured EHR data into representations that can be used as inputs to ML algorithm" (Tang et al., 2020). Examples of how this preprocessing pipeline works include identification of time varying data; filtering and collapsing rare, duplicate, or uninformative data; identification of frequent versus infrequent measures; missing data imputation; and automatically reconciling heterogenous data types. The goal with a pipeline such as FIDDLE is to enable researchers to obtain EHR data beyond what is easy or convenient.
One immediate application of ML to knowledge discovery is improving EHR data abstraction. Whereas manual chart review is cumbersome, prone to errors, and time consuming, ML has been deployed to automate the process for both structured and unstructured data. Hu et al. (2016) explored the use of six automated approaches for identifying postoperative complications among surgical patients using structured EHR data, including diagnostic codes, microbiology tests, antibiotic administration, and other labs. The performance of each of these approaches was compared against manual chart review, which was treated as the gold standard, and all approaches performed better than chance alone (area under the curve exceeding 0.8 for all complications). In Park et al. (2020), the authors contrasted three ML algorithms for identifying hospital-acquired catheter-associated urinary tract infections from EHR data. Similar to Hu, they used structured data from the EHR, supplemented with ancillary data from nursing, and constructed a gold standard via manual chart review. Also similar to Hu, the authors observed that the algorithms performed better than chance (area under the curve exceeding 0.75 for all algorithms) and noted the feasibility of ML for data mining hospital-acquired catheter-associated urinary tract infections.
Data need not be limited to the healthcare enterprise for this application. Researchers have linked publicly available data on the built environment, neighborhood characteristics, and housing attributes to the EHR to explore in home environmental exposures to asthma (Bozigar et al., 2022). Using an ensemble approach, four allergen prediction models were constructed to discover the presence of cockroaches, rodents, mold, and carpets. The gold standard in this analysis was an asthma flowsheet in the EHR that was only occasionally completed by the healthcare workers. The algorithms performed modestly (area under the curve between 0.55 and 0.65) but demonstrated the possibility of supplementing existing clinical data with public data to identify harmful exposures not otherwise captured in the EHR.
Data also need not be structured to automatically derive meaning; this was covered in a previous blog post.
Cite: Goldstein ND. Machine learning in electronic health record research. Dec 28, 2022. DOI: 10.17918/goldsteinepi.